Taxonomy Phylogeny
Taxonomies group organisms according to phenotype, while phylogenetic systems groups organisms according to shared evolutionary heritage.
Taxonomy
Taxonomies aim to group organisms according to shared characteristics against the background of biological diversity.
The major subdivisions employed in general taxonomies are: Domain, Kingdom, Phylum, Class, Order, Family, Genus, and Species. The mnemonic "Dashing King Philip Came Over From Greater Spain" applies.
These subdivisions may be further subdivided. It is common for bacteria to be subdivided into Divisions and further subdivided into Orders. The advent of genomics and examination of 16s ribosomal RNA has led to modifications in the classification of the prokaryotes, with the older nomenclatures revised into new classifications of Phyla.
Numerical Taxonomy: a common approach to phenetic taxonomy, which employs a number of phenotypic characteristics to generate similarity coefficients that may be mapped in dendrograms. Groupings based on numerical taxonomy may or may not correlate with evolutionary relationships.
Labels: Class, Domain, Family, Genus, Kingdom, Order, Phylum, species, taxonomy
Phylogenetics
Phylogenetic separation into evolutionary relationships (clades), based on comparison of genomes is likely to supplant phenotypical (phenetic) taxonomies of the prokaryotes.
Phenetic system: groupings of organisms based on mutual similarity of phenotypic (physical and chemical) characteristics. Phenetic groupings may or may not correlate with evolutionary relationships.
Phylogenetic system: groups organisms based on shared evolutionary heritage. DNA and RNA sequencing techniques are considered to give the most meaningful phylogenies.
Monophyletic taxon or clade: an accurate grouping of only (opp. polyphyletic) and all (opp. paraphyletic) descendents of a shared common ancestor. A monopyletic group is genetically homogeneous and reflects evolutionary relationships.
Paraphyletic taxon or clade: a monophyletic group that excludes one or more discrete groups descended from the most recent common ancestral species of the entire group. Other descendent species of the most recent common ancestor have been excluded from the paraphyletic taxon, usually because of morphologic distinctiveness.
Polyphyletic taxon: opposite to monophyletic taxon: A polyphyletic group is mistakenly or improperly erected on the basis of homoplasy.—characteristics that have arisen despite not sharing a common ancestor. Homoplasy arises because of convergent evolution, parallelism, evolutionary reversals, horizontal gene transfer, or gene duplications. Polyphyletic taxa are genetically heterogeneous because members do not share a common ancestor.
Labels: monophyletic, paraphyletic, phenetic, phylogenetics, polyphyletic, taxon
homology homoplasy
Homology refers to common ancestry of two or more genes or gene products that have evolved from the same feature in the last common ancestor of the species. When applied to nucleotide or protein sequences, homology refers specifically to this relationship due to descent from a common ancestral sequence.
Homologous features are derived through the same evolutionary or phylogenetic origin, yet do not necessarily share the same function or structure. Homologous genes share an arbitrary threshold level of similarity determined by alignment of matching bases. Such homology is an important organizing principle for genomic studies because structural and functional similarities tend to change together along the structure of homology relationships. It may be difficult to determine that genes share a common ancestor when their structure has been modified through descent. Functional genes often display homology across species because evolutionary tinkering that destroys essential functions could be detrimental to the survival of the organism, and hence to transmission of the altered gene.
Homology is a qualitative term (+/-), while similarity is the corresponding quantitative term in that we refer to degrees of similarity. Homology is distinguished from homoplasy, which refers to the similarity between structures that is due to common ancestry. In cladistics, both homology and homoplasy are determined a posteriori with reference to a particular phylogeny that maximizes homology and minimizes homoplasy. The amount of homology between species may be used to determine evolutionary relationships and degrees of divergence.
Homeobox genes provide an example of functional conservation that is misapplied by creationists to refute biological evolution. Hox genes control early development and necessarily show high degrees of homology across species.
Labels: cladistics, homeobox genes, homology, homoplasy, Hox genes, phylogenetics
History of taxonomic concepts
History of taxonomic concepts:
Linnaeus, 1735 – 2 Kingdoms – Animalia, Vegetabilia
Haeckel, 1866 – 3 Kingdoms – Protista, Plantae, Animalia. Image Haeckel's tree of life.
Chatton, 1937 – 2 Empires – Prokaryota, Eukaryota
Copeland, 1956 – 4 Kingdoms – Monera (prokaryotes), Protoctista, Plantae, Animalia
Whittaker, 1969 – Monera (prokaryotes), Fungi, Protista, Plantae, Animalia
Woese et al, 1977 – 6 Kingdom – Eubacteria, Archaea, Protista, Fungi, Plantae, Animalia
Woese and Fox, 1999 – 3 Domain system: Eubacteria, Archaea, and Eukaryotes.
Table Comparisons of Eubacteria, Archaea, and Eukaryotes - The Three Domains View Quicktime Movie - Genetic Data movie of phylogram construction - image cladogram - image Tree of Life— Lateral Gene Transfer Diagram - image uprooted tree - image 16S ribosomal RNA - image The "Shrub of Life" - image A comparison of key characteristics from the three domains of life - enlarged - image Whale phylogeny - Genomics Animations and Images - Proteins & Proteomics - Animations and Images – Evolution and Phylogenetics - Animations and Images - Human Evolution - Animations and Images - Genetics of Development - Animations and Images – Cell Biology & Cancer - Animations and Images - Neurobiology - Animations and Images - Biology of Sex & Gender - Animations and Images - Genetically modified organisms - Animations and Images - Biodiversity - Animations and Images – Microbial Diversity – Animations and Images – Emerging Infectious Diseases - Animations and Images – HIV & AIDS - Animations and Images :
Labels: Chatton, Copeland, Fox, Haeckel, history, Linnaeus, Whittaker, Woese
Archaea Eocyte Tree

According to the Tree of Life Web Project, two alternative views on the relationship of the major lineages (omitting viruses) are currently regarded as viable (right - click to enlarge image).
Labels: Archaea, Eocyte, Eubacteria, Eukaryotes
Bacteria
Taxonomy of the bacteria was historically based on phenotypical physical and chemical characteristics – phenetic taxonomy. According to "Bergey's Manual of Systematic Bacteriology", all bacteria can be classified into four divisions, or phyla according to the constituents of their cell walls.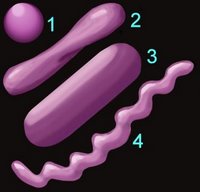 Each division was further subdivided into sections according to :
Each division was further subdivided into sections according to :
1. Reaction of the cell wall to the Gram stain due to thick layer of peptidoglycan (Gram +ve) or thin layer of peptidoglycan (Gram -ve). The cell walls of Archaeobacteria contain no peptidoglycan.
2. Shape (left) – cocci (1), pleomorphic (2), bacillus (3), helical (4)
3. Arrangement of cells
4. Oxygen requirement – aerobic, anaerobic, or facultative anaerobe
5. Motility – flagellated, non-motile
6. Specific nutritional requirements
7. Metabolic properties – autotrophic (chemical or photosynthetic), heterotrophic
The advent of genomics and examination of 16s ribosomal RNA has led to modifications in the classification of the prokaryotes, with the older nomenclatures revised into new classifications of Phyla. (diagram at bottom) Woese and Fox proposed the Three Domain system: Eubacteria, Archaea, and Eukaryotes. When Woese and Fox proposed the 3 Domain system, the term 'Urkaryotes' was proposed for ancestors of eukaryotes prior to their endosymbiotic acquisition of mitochondria and chloroplasts from prokaryotes.
"Misunderstanding the Bacteriological Code"
The Three Domain system is increasingly accepted. Free Full Text Article : Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Woese CR, Fox GE. Proc Natl Acad Sci U S A. 1977 Nov;74(11):5088-90.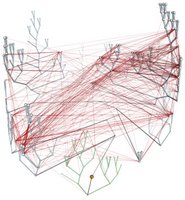 Left - click to enlarge image: Horizontal gene transfer - gene swapping - has blurred the evolutionary relationships (image) of prokaryotes, and continues to provide a mechanism for the sharing of antibiotic resistance between bacteria. See: Trees, vines and nets: Microbial evolution changes its face.
Left - click to enlarge image: Horizontal gene transfer - gene swapping - has blurred the evolutionary relationships (image) of prokaryotes, and continues to provide a mechanism for the sharing of antibiotic resistance between bacteria. See: Trees, vines and nets: Microbial evolution changes its face.
Phylogenetic separation into evolutionary relationships (clades), based on comparison of genomes is likely to supplant phenotypical (phenetic) taxonomies of the prokaryotes.(nomenclature)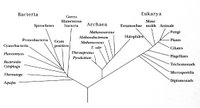
Right - click to enlarge image: Based on Woese's proposed scheme based on the 16s subunit of ribosomal RNA, which appears to better illustrate evolutionary relationships within the 3 domains.
Labels: 16s rRNA, 3 Domain, Archaeobacteria, bacteria, Eubacteria, Fox, gram, taxonomy, Woese
Cambrian explosion
"The key element in deciphering the Cambrian explosion (11) is to integrate the expanding insights of molecular phylogeny (12) and developmental biology with the totality of paleontological evidence, including the Ediacaran assemblages. Somewhere, and this is the tricky point, in the Ediacaran assemblages are animals that may throw particular light on key transitions. Of these, the most significant are those between sponges and diploblasts, cnidarians and triploblasts, as well as the early evolution of the three superclades of triploblast (deuterostomes, ecdysozoans, and lophotrochozoans) (Fig. 1). The overall framework of early metazoan evolution comes from molecular data, but they cannot provide insights into the anatomical changes and associated changes in ecology that accompanied the emergence of bodyplans during the Cambrian explosion. The fossil record provides, therefore, a unique historical perspective."
Simon Conway Morris The Cambrian "explosion": Slow-fuse or megatonnage? PNAS April 25, 2000 vol. 97 no. 9 4426-4429
Labels: Cambrian explosion
Endosymbiotic transfer events
Right - click to enlarge image: Proposed endosymbiotic transfer events between the three Domains and the six Kingdoms of Life.
Both the Eubacteria and Archaea are prokaryotes, while animals, fungi, plants, and protists are eukaryotes.
The yellow asterisk * indicates the last universal common ancestor (LUCA), or universal cenancestor, which is hypothesized as being at the ancestral root of all living organisms. Not the earliest or simplest living organism, and not necessarily the sole example of its type, this organism possessed the genetic material that diverged (about 3.5 Ga) into all current living organisms. A number of terms are employed to refer to the universal cenancestor – last universal ancestor (LUA), last common ancestor (LCA), or last universal common ancestor (LUCA).
Labels: Archaea, endosymbiotic transfer events, Eubacteria, Eukaryotes, LUCA, serial endosymbiosis








































